Here are two papers that may be useful for researchers in psychology, linguistics, and cognitive science:
Shravan Vasishth and Bruno Nicenboim. Statistical methods for linguistic research: Foundational Ideas - Part I. Language and Linguistics Compass, 2016. In Press.
PDF: http://bit.ly/VasNicPart1
Code: http://bit.ly/VasNicPart1Code
Bruno Nicenboim and Shravan Vasishth. Statistical methods for linguistics research: Foundational Ideas - Part II. Language and Linguistics Compass, 2016. In Press.
PDF: http://bit.ly/NicVasPart2
Code: http://bit.ly/NicVasPart2Code
This blog is a repository of cool things relating to statistical computing, simulation and stochastic modeling.
Search
Tuesday, August 02, 2016
Wednesday, April 27, 2016
A simple proof that the p-value distribution is uniform when the null hypothesis is true
[Scroll to graphic below if math doesn't render for you]
Thanks to Mark Andrews for correcting some crucial typos (I hope I got it right this time!).
Thanks also to Andrew Gelman for pointing out that the proof below holds only when the null hypothesis is a point null $H_0: \mu = 0$, and the dependent measure is continuous, such as reading time in milliseconds, or EEG responses.
Someone asked this question in my linear modeling class: why is it that the p-value has a uniform distribution when the null hypothesis is true? The proof is remarkably simple (and is called the probability integral transform).
First, notice that when a random variable Z comes from a $Uniform(0,1)$ distribution, then the probability that $Z$ is less than (or equal to) some value $z$ is exactly $z$: $P(Z\leq z)=z$.
Next, we prove the following proposition:
Proposition:
If a random variable $Z=F(T)$, then $Z \sim Uniform(0,1)$.
Note here that the p-value is a random variable, call it $Z$. The p-value is computed by calculating the probability of seeing a t-statistic or something more extreme under the null hypothesis. The t-statistic comes from a random variable $T$ that is a transformation of the random variable $\bar{X}$: $T=(\bar{X}-\mu)/(\sigma/\sqrt{n})$. This random variable T has a CDF $F$.
So, if we can prove the above proposition, we have shown that the p-value's distribution under the null hypothesis is $Uniform(0,1)$.
Proof:
Let $Z=F(T)$.
$P(Z\leq z) = P(F(T)\leq z) = P(F^{-1} F(T) \leq F^{-1}(z) )
= P(T \leq F^{-1} (z) )
= F(F^{-1}(z))= z$.
Since $P(Z\leq z)=z$, Z is uniformly distributed, that is, Uniform(0,1).
A screengrab in case the above doesn't render:
Thanks to Mark Andrews for correcting some crucial typos (I hope I got it right this time!).
Thanks also to Andrew Gelman for pointing out that the proof below holds only when the null hypothesis is a point null $H_0: \mu = 0$, and the dependent measure is continuous, such as reading time in milliseconds, or EEG responses.
Someone asked this question in my linear modeling class: why is it that the p-value has a uniform distribution when the null hypothesis is true? The proof is remarkably simple (and is called the probability integral transform).
First, notice that when a random variable Z comes from a $Uniform(0,1)$ distribution, then the probability that $Z$ is less than (or equal to) some value $z$ is exactly $z$: $P(Z\leq z)=z$.
Next, we prove the following proposition:
Proposition:
If a random variable $Z=F(T)$, then $Z \sim Uniform(0,1)$.
Note here that the p-value is a random variable, call it $Z$. The p-value is computed by calculating the probability of seeing a t-statistic or something more extreme under the null hypothesis. The t-statistic comes from a random variable $T$ that is a transformation of the random variable $\bar{X}$: $T=(\bar{X}-\mu)/(\sigma/\sqrt{n})$. This random variable T has a CDF $F$.
So, if we can prove the above proposition, we have shown that the p-value's distribution under the null hypothesis is $Uniform(0,1)$.
Proof:
Let $Z=F(T)$.
$P(Z\leq z) = P(F(T)\leq z) = P(F^{-1} F(T) \leq F^{-1}(z) )
= P(T \leq F^{-1} (z) )
= F(F^{-1}(z))= z$.
Since $P(Z\leq z)=z$, Z is uniformly distributed, that is, Uniform(0,1).
A screengrab in case the above doesn't render:
Sunday, January 17, 2016
Automating R exercises and exams using the exams package
It's a pain to design statistics exercises each semester, and because students from previous share old exercises with the new incoming students, it's hard to design simple exercises that students haven't already seen the answers to. On top of that, some students try to cheat during the exam by looking over the shoulder of their neighbors. Homework exercises almost always involve collaboration even if you prohibit it.
It turns out that you can automate the generation of fixed-format exercises (with different numerical answers being required each time). You can also randomly select questions from a question bank you create yourself. And you can even create a unique question paper for each student in an exam, to make cheating between neighbors essentially impossible (even if they copy the correct answer to question 2 from a neighbor, they end up answering the wrong question on their own paper).
All this magic is made possible by the exams package in R. The documentation is of course comprehensive and there is a journal article explaining everything:
Here is a quick example to get people started on designing their own customized, automated exams. In my example below, there are several files you need.
1. The template files for your exam (what your exam or homework sheet will look like), and the solutions file. I provide two example files: test.tex and solutiontest.tex
2. The exercises or exam questions themselves: I provide two as examples. The first file is called pnorm1.Rnw. It's an Sweave file, and it contains the code for generating a random problem and for generating its solution. The code should be self-explanatory. The second file is called sesamplesize1multiplechoice.Rnw and has a multiple choice question.
3. The exam generating R code file: The code is commented and self-explanatory. It will generate the exercises, randomize the order of presentation (if there are two or more exercises), and generate a solutions file. The output will be a single or multiple exam papers (depending on how many versions you wanted generated), and the solutions file(s). Notice the cool thing that even in my example, with only one question, the two versions of the exams have different numbers, so two people cannot collaborate and consult each other and just write down one answer. Each student could in principle be given a unique set of exercises, although it would be a lot of work to grade it if you do it manually.
Here is the exam generating code:
Save from the gists given above (a) the test.tex and solutiontest.tex files, (b) the Rnw files containing the exercise (pnorm1.Rnw, and sesamplesize1multiplechoice.Rnw), and (c) the exam generating code (ExampleExamCode.R). Put all of these into your working directory, say ExampleExam. Then run the R code, and be amazed.
If something is broken in my example, please let me know.
Shuffling questions: If you want to reorder the questions in each run of the R code, just change myexamlist to sample(myexamlist) in the call below that appears in the file ExampleExamCode.R:
It turns out that you can automate the generation of fixed-format exercises (with different numerical answers being required each time). You can also randomly select questions from a question bank you create yourself. And you can even create a unique question paper for each student in an exam, to make cheating between neighbors essentially impossible (even if they copy the correct answer to question 2 from a neighbor, they end up answering the wrong question on their own paper).
All this magic is made possible by the exams package in R. The documentation is of course comprehensive and there is a journal article explaining everything:
Achim Zeileis, Nikolaus Umlauf, Friedrich Leisch (2014). Flexible Generation of E-Learning Exams in R: Moodle Quizzes, OLAT Assessments, and Beyond. Journal of Statistical Software 58(1), 1-36. URL http://www.jstatsoft.org/v58/i01/.I also use this package to deliver auto-graded exercises to students over datacamp.com. See here for the course I teach, and here for the datacamp exercises.
Here is a quick example to get people started on designing their own customized, automated exams. In my example below, there are several files you need.
1. The template files for your exam (what your exam or homework sheet will look like), and the solutions file. I provide two example files: test.tex and solutiontest.tex
2. The exercises or exam questions themselves: I provide two as examples. The first file is called pnorm1.Rnw. It's an Sweave file, and it contains the code for generating a random problem and for generating its solution. The code should be self-explanatory. The second file is called sesamplesize1multiplechoice.Rnw and has a multiple choice question.
3. The exam generating R code file: The code is commented and self-explanatory. It will generate the exercises, randomize the order of presentation (if there are two or more exercises), and generate a solutions file. The output will be a single or multiple exam papers (depending on how many versions you wanted generated), and the solutions file(s). Notice the cool thing that even in my example, with only one question, the two versions of the exams have different numbers, so two people cannot collaborate and consult each other and just write down one answer. Each student could in principle be given a unique set of exercises, although it would be a lot of work to grade it if you do it manually.
Here is the exam generating code:
Save from the gists given above (a) the test.tex and solutiontest.tex files, (b) the Rnw files containing the exercise (pnorm1.Rnw, and sesamplesize1multiplechoice.Rnw), and (c) the exam generating code (ExampleExamCode.R). Put all of these into your working directory, say ExampleExam. Then run the R code, and be amazed.
If something is broken in my example, please let me know.
Shuffling questions: If you want to reorder the questions in each run of the R code, just change myexamlist to sample(myexamlist) in the call below that appears in the file ExampleExamCode.R:
sol <- exams(sample(myexamlist), n = num.versions,
dir = odir, template = c("test", "solutiontest"),
nsamp=1,
header = list(ID = getID, Date = Sys.Date()))
Wednesday, January 06, 2016
My MSc thesis: A meta-analysis of relative clause processing in Mandarin Chinese using bias modelling
Here is my MSc thesis, which was submitted to the University of Sheffield in September 2015.
The pdf is here.
Title: A Meta-analysis of Relative Clause Processing in Mandarin Chinese using Bias Modelling
Abstract
The reading difficulty associated with Chinese relative clauses presents an important empirical problem for psycholinguistic research on sentence comprehension processes. Some studies show that object relatives are easier to process than subject relatives, while others show the opposite pattern. If Chinese has an object relative advantage, this has important implications for theories of reading comprehension. In order to clarify the facts about Chinese, we carried out a Bayesian random-effects meta-analysis using 15 published studies; this analysis showed that the posterior probability of a subject relative advantage is approximately $0.77$ (mean $16$, 95% credible intervals $-29$ and $61$ ms). Because the studies had significant biases, it is possible that they may have confounded the results. Bias modelling is a potentially important tool in such situations because it uses expert opinion to incorporate the biases in the model. As a proof of concept, we first identified biases in five of the fifteen studies, and elicited priors on these using the SHELF framework. Then we fitted a random-effects meta-analysis, including priors on biases. This analysis showed a stronger posterior probability ($0.96$) of a subject relative advantage compared to the standard random-effects meta-analysis (mean $33$, credible intervals $-4$ and $71$).
The pdf is here.
Title: A Meta-analysis of Relative Clause Processing in Mandarin Chinese using Bias Modelling
Abstract
The reading difficulty associated with Chinese relative clauses presents an important empirical problem for psycholinguistic research on sentence comprehension processes. Some studies show that object relatives are easier to process than subject relatives, while others show the opposite pattern. If Chinese has an object relative advantage, this has important implications for theories of reading comprehension. In order to clarify the facts about Chinese, we carried out a Bayesian random-effects meta-analysis using 15 published studies; this analysis showed that the posterior probability of a subject relative advantage is approximately $0.77$ (mean $16$, 95% credible intervals $-29$ and $61$ ms). Because the studies had significant biases, it is possible that they may have confounded the results. Bias modelling is a potentially important tool in such situations because it uses expert opinion to incorporate the biases in the model. As a proof of concept, we first identified biases in five of the fifteen studies, and elicited priors on these using the SHELF framework. Then we fitted a random-effects meta-analysis, including priors on biases. This analysis showed a stronger posterior probability ($0.96$) of a subject relative advantage compared to the standard random-effects meta-analysis (mean $33$, credible intervals $-4$ and $71$).
Saturday, December 19, 2015
Best statistics-related comment ever from a reviewer
This is the most interesting comment I have ever received from CUNY conference reviewing. It nicely illustrates the state of our understanding of statistical theory in psycholinguistics:
"I had no idea how many subjects each study used. Were just one or two people
used? ... Generally, I wasn't given enough data to determine my confidence in the provided t-values (which depends on the degrees of freedom involved)."
"I had no idea how many subjects each study used. Were just one or two people
used? ... Generally, I wasn't given enough data to determine my confidence in the provided t-values (which depends on the degrees of freedom involved)."
Thursday, August 27, 2015
Five thirty-eight provides a brand new definition of the p-value
The Five-Thirty Eight blog provides a brand new definition of the p-value:
http://fivethirtyeight.com/datalab/psychology-is-starting-to-deal-with-its-replication-problem/?ex_cid=538twitter
"A p-value is simply the probability of getting a result at least as extreme as the one you saw if your hypothesis is false."
I thought this blog was run by Nate Silver, a statistician?
http://fivethirtyeight.com/datalab/psychology-is-starting-to-deal-with-its-replication-problem/?ex_cid=538twitter
"A p-value is simply the probability of getting a result at least as extreme as the one you saw if your hypothesis is false."
I thought this blog was run by Nate Silver, a statistician?
Observed vs True Statistical Power, and the power inflation index
People (including me) routinely estimate statistical power for future studies using a pilot study's data or a previously published study's data (or perhaps using the predictions from a computational model, such as Engelmann et al 2015).
Indeed, the author of the Replicability Index has been using observed power to determine the replicability of journal articles. His observed power estimates are HUGE (in the range of 0.75) and seem totally implausible to me, given the fact that I can hardly ever replicate my studies.
This got me thinking: Gelman and Carlin have shown that when power is low, Type M error will be high. That is, the observed effects will tend to be highly exaggerated. The issue with Type M error is easy to visualize.
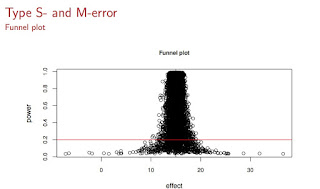
Suppose that a particular study has standard error 46, and sample size 37; this implies that standard deviation is $46\times \sqrt{37}= 279$. These are representative numbers from psycholinguistic studies. Suppose also that we know that the true effect (the absolute value, say on the millisecond scale for a reading study---thanks to Fred Hasselman) is D=15. Then, we can compute Type S and Type M errors for replications of this particular study by repeatedly sampling from the true distribution.
We can visualize the exaggerated effects under low power as follows (see below): On the x-axis you see the effect magnitudes, and on the y-axis is power. The red line is the power line of 0.20, which based on my own attempts at replicating my own studies (and mostly failing), I estimate to be an upper bound of the power of experiments in psycholinguistics (this is an upper bound, I think a more common value will be closer to 0.05). All those dots below the red line are exaggerated estimates in a low power situation, and if you were to use any of those points to estimate observed power, you would get a wildly optimistic power estimate which has no bearing with reality.
What does this fact about Type M error imply for Replicability Index's calculations? It implies that if power is in fact very low, and if journals are publishing larger-than-true effect sizes (and we know that they have an incentive to do so, because editors and reviewers mistakenly think that lower p-values, i.e., bigger absolute t-values, give stronger evidence for the specific alternative hypothesis of interest), then Replicability Index is possibly hugely overestimating power, and therefore hugely overestimating replicability of results.
I came up with the idea of framing this overestimation in terms of Type M error by defining something called a power inflation index. Here is how it works. For different "true" power levels, we repeatedly sample data, and compute observed power each time. Then, for each "true" power level, we can compute the ratio of the observed power to the true power in each case. The mean of this ratio is the power inflation index, and the 95% confidence interval around it gives us an indication (sorry Richard Morey! I know I am abusing the meaning of CI here and treating it like a credible interval!) of how badly we could overestimate power from a small sample study.
Here is the code for simulating and visualizing the power inflation index:
And here is the visualization:
What we see here is that if true power is as low as 0.05 (and we can never know that it is not since we never know the true effect size!), then using observed power can lead to gross overestimates by a factor of approximately 10! So, if Replicability Index reports an observed power of 0.75, what he might actually be looking at is an inflated estimate where true power is 0.08.
In summary, we can never know true power, and if we are estimating it using observed power conditional on true power being extremely low, we are likely to hugely overestimate power.
One way to test my claim is to actually try to replicate the studies that Replicability Index predicts has high replicability. My prediction is that his estimates will be wild overestimates and most studies will not replicate.
Postscript
A further thing that worries me about Replicability Index is his sloppy definitions of statistical terms. Here is how he defines power:
"Power is defined as the long-run probability of obtaining significant results in a series of exact replication studies. For example, 50% power means that a set of 100 studies is expected to produce 50 significant results and 50 non-significant results."
[Thanks to Karthik Durvasula for correcting my statement below!]
By not defining power of a test of a null hypothesis $H_0: \mu=k$, as the probability of rejecting the null hypothesis (as a function of different alternative $\mu$ such that $\mu\neq k$) when it is false, what this definition literally means is that if I sample from any distribution, including one where $\mu=0$, the probability of obtaining a significant result under repeated sampling is the power. Which of course is completely false.
Post-Post Script
Replicability Index points out in a tweet that his post-hoc power estimation corrects for inflation. But post-hoc power corrected for inflation requires knowledge of the true power, which is what we are trying to get at in the first place. How do you deflate "observed" power when you don't know what the true power is? Maybe I am missing something.
Indeed, the author of the Replicability Index has been using observed power to determine the replicability of journal articles. His observed power estimates are HUGE (in the range of 0.75) and seem totally implausible to me, given the fact that I can hardly ever replicate my studies.
This got me thinking: Gelman and Carlin have shown that when power is low, Type M error will be high. That is, the observed effects will tend to be highly exaggerated. The issue with Type M error is easy to visualize.
Suppose that a particular study has standard error 46, and sample size 37; this implies that standard deviation is $46\times \sqrt{37}= 279$. These are representative numbers from psycholinguistic studies. Suppose also that we know that the true effect (the absolute value, say on the millisecond scale for a reading study---thanks to Fred Hasselman) is D=15. Then, we can compute Type S and Type M errors for replications of this particular study by repeatedly sampling from the true distribution.
We can visualize the exaggerated effects under low power as follows (see below): On the x-axis you see the effect magnitudes, and on the y-axis is power. The red line is the power line of 0.20, which based on my own attempts at replicating my own studies (and mostly failing), I estimate to be an upper bound of the power of experiments in psycholinguistics (this is an upper bound, I think a more common value will be closer to 0.05). All those dots below the red line are exaggerated estimates in a low power situation, and if you were to use any of those points to estimate observed power, you would get a wildly optimistic power estimate which has no bearing with reality.
What does this fact about Type M error imply for Replicability Index's calculations? It implies that if power is in fact very low, and if journals are publishing larger-than-true effect sizes (and we know that they have an incentive to do so, because editors and reviewers mistakenly think that lower p-values, i.e., bigger absolute t-values, give stronger evidence for the specific alternative hypothesis of interest), then Replicability Index is possibly hugely overestimating power, and therefore hugely overestimating replicability of results.
I came up with the idea of framing this overestimation in terms of Type M error by defining something called a power inflation index. Here is how it works. For different "true" power levels, we repeatedly sample data, and compute observed power each time. Then, for each "true" power level, we can compute the ratio of the observed power to the true power in each case. The mean of this ratio is the power inflation index, and the 95% confidence interval around it gives us an indication (sorry Richard Morey! I know I am abusing the meaning of CI here and treating it like a credible interval!) of how badly we could overestimate power from a small sample study.
Here is the code for simulating and visualizing the power inflation index:
And here is the visualization:
What we see here is that if true power is as low as 0.05 (and we can never know that it is not since we never know the true effect size!), then using observed power can lead to gross overestimates by a factor of approximately 10! So, if Replicability Index reports an observed power of 0.75, what he might actually be looking at is an inflated estimate where true power is 0.08.
In summary, we can never know true power, and if we are estimating it using observed power conditional on true power being extremely low, we are likely to hugely overestimate power.
One way to test my claim is to actually try to replicate the studies that Replicability Index predicts has high replicability. My prediction is that his estimates will be wild overestimates and most studies will not replicate.
Postscript
A further thing that worries me about Replicability Index is his sloppy definitions of statistical terms. Here is how he defines power:
"Power is defined as the long-run probability of obtaining significant results in a series of exact replication studies. For example, 50% power means that a set of 100 studies is expected to produce 50 significant results and 50 non-significant results."
[Thanks to Karthik Durvasula for correcting my statement below!]
By not defining power of a test of a null hypothesis $H_0: \mu=k$, as the probability of rejecting the null hypothesis (as a function of different alternative $\mu$ such that $\mu\neq k$) when it is false, what this definition literally means is that if I sample from any distribution, including one where $\mu=0$, the probability of obtaining a significant result under repeated sampling is the power. Which of course is completely false.
Post-Post Script
Replicability Index points out in a tweet that his post-hoc power estimation corrects for inflation. But post-hoc power corrected for inflation requires knowledge of the true power, which is what we are trying to get at in the first place. How do you deflate "observed" power when you don't know what the true power is? Maybe I am missing something.
Monday, August 17, 2015
Some reflections on teaching frequentist statistics at ESSLLI 2015
I spent the last two weeks teaching frequentist and Bayesian statistics at the European Summer School in Logic, Language, and Information (ESSLLI) in Barcelona, at the beautiful and centrally located Pompeu Fabra University. The course web page for the first week is here, and the web page for the second course is here.
All materials for the first week are available on github, see here.
The frequentist course went well, but the Bayesian course was a bit unsatisfactory; perhaps my greater experience in teaching the frequentist stuff played a role (I have only taught Bayes for three years). I've been writing and rewriting my slides and notes for frequentist methods since 2002, and it is only now that I can present the basic ideas in five 90 minute lectures; with Bayes, the presentation is more involved and I need to plan more carefully, interspersing on-the-spot exercises to solidify ideas. I will comment on the Bayesian Data Analysis course in a subsequent post.
The first week (five 90 minute lectures) covered the basic concepts in frequentist methods. The audience was amazing; I wish I always had students like these in my classes. They were attentive, and anticipated each subsequent development. This was the typical ESSLLI crowd, and this is why teaching at ESSLLI is so satisfying. There were also several senior scientists in the class, so hopefully they will go back and correct the misunderstandings among their students about what all this Null Hypothesis Significance Testing stuff gives you (short answer: it answers *a* question very well, but it's the wrong question, nothing that is relevant to your research question).
I won't try to summarize my course, because the web page is online and you can also do exercises on datacamp to check your understanding of statistics (see here). You get immediate feedback on your attempts.
Stepping away from the technical details, I tried to make three broad points:
First, I spent a lot of time trying to clarify what a p-value is and isn't, focusing particularly on the issue of Type S and Type M errors, which Gelman and Carlin have discussed in their excellent paper.
Here is the way that I visualized the problems of Type S and Type M errors:
What we see here is repeated samples from a Normal distribution with true mean 15 and a typical standard deviation seen in psycholinguistic studies (see slide 42 of my slides for lecture2). The horizontal red line marks the 20% power line; most psycholinguistic studies fall below that line in terms of power. The dramatic consequence of this low power is the hugely exaggerated effects (which tend to get published in major journals because they also have low p-values) and the remarkable proportion of cases where the sample mean is on the wrong side of the true value 15. So, you are roughly equally likely to get a significant effect with a sample mean smaller to much smaller than the true mean, or larger and much larger than the true mean. With low power, regardless of whether you get a significant result or not, if power is low, and it is in most studies I see in journals, you are just farting in a puddle.
It is worth repeating this: once one considers Type S and Type M errors, even statistically significant results become irrelevant, if power is low. It seems like these ideas are forever going to be beyond the comprehension of researchers in linguistics and psychology, who are trained to make binary decisions based on p-values, weirdly accepting the null if p is greater than 0.05 and, just as weirdly, accepting their favored alternative if p is less than 0.05. The p-value is a truly interesting animal; it seems that a recent survey of some 400 Spanish psychologists found that, despite their being active in the field for quite a few years on average, they had close to zero understanding of what a p-value gives you. Editors of top journals in psychology routinely favor lower p-values, because they mistakenly think this makes "the result" more convincing; "the result" is the favored alternative. So even seasoned psychologists (and I won't even get started with linguists, because we are much, much worse), with decades of experience behind them, often have no idea what the p-value actually tells you.
A remarkable misunderstanding regarding p-values is the claim that it tells you whether the effect was "by chance". Here is an example from Replication Index's blog:
"The Test of Insufficient Variance (TIVA) shows that the variance in z-scores is less than 1, but the probability of this event to occur by chance is 10%, Var(z) = .63, Chi-square (df = 11) = 17.43, p = .096."
Here is another example from a self-published manuscript by Daniel Ezra Johnson:
"If we perform a likelihood-ratio test, comparing the model with gender to a null model with no predictors, we get a p-value of 0.0035. This implies that it is very unlikely that the observed gender difference is due to chance."
One might think that the above examples are not peer-reviewed, and that peer review would catch such mistakes. But even people explaining p-values in publications are unable to understand that this is completely false. An example is Keith Johnson's textbook, Quantitative Methods in Linguistics, which repeatedly talks about "reliable effects" and effects which are and are not due to chance. It is no wonder that the poor psychologist/linguist thinks, ok, if the p-value is telling me the probability that the effect is due to chance, and if the p-value is low, then the effect is not due to chance and the effect must be true. The mistake here is that the p-value is telling you the probability of the result being "due to chance" conditional on the null hypothesis being true. It's better to explain the p-value as the probability of getting the statistic (e.g., t-value) or something more extreme, under the assumption that the null hypothesis is true. People seem to drop the italicized part and this starts to propagate the misunderstanding for future generations. To repeat, the p-value is a conditional probability, but most people interpret it as an unconditional probability because they drop the phrase "under the null hypothesis" and truncate the statement to be about effects being due to chance.
Another bizarre thing I have repeatedly seen is misinterpreting the p-value as Type I error. Type I error is fixed at a particular value (0.05) before you run the experiment, and is the probability of your incorrectly rejecting the null when it's true, under repeated sampling. The p-value is what you get from your single experiment and is the conditional probability of your getting the statistic you got or something more extreme, assuming that the null is true. Even this point is beyond comprehension for psychologists (and of course linguists). Here is a bunch of psychologists explaining in an article why a p=0.000 should not be reported as an exact value:
The p-value is the probability of committing a Type I error, eh? It is truly embarrassing that people who are teaching this stuff have distorted the meaning of the p-value so drastically and just keep propagating the error. I should mention though that this paper I am citing appeared in Frontiers, which I am beginning to question as a worthwhile publication venue. Who did the peer review on this paper and why did they not catch this basic mistake?
Even Fisher (p. 16 of The Design of Experiments, Second Edition, 1937) didn't buy the p-value; he is advocating for replicability as the real decisive tool:
"It is usual and convenient for experimenters to take-5 per cent. as a standard level of significance, in the sense that they are prepared to ignore all results which fail to reach this standard, and, by this means, to eliminate from further discussion the greater part of the fluctuations which chance causes have introduced into their experimental results. No such selection can eliminate the whole of the possible effects of chance. coincidence, and if we accept this convenient convention, and agree that an event which would occur by chance only once in 70 trials is decidedly" significant," in the statistical sense, we thereby admit that no isolated experiment, however significant in itself, can suffice for the experimental demonstration of any natural phenomenon; for the "one chance in a million" will undoubtedly occur, with no less and no more than its appropriate frequency, however surprised we may be that it should occur to us. In order to assert that a natural phenomenon is experimentally demonstrable we need, not an isolated record, but a reliable method of procedure. In relation to the test of significance, we may say that a phenomenon is experimentally demonstrable when we know how to conduct an experiment which will rarely fail to give us a statistically significant result."
Second, I tried to clarify what a 95% confidence interval is and isn't. At least a couple of students had a hard time accepting that the 95% CI refers to the procedure and not that the true $\mu$ lies within one specific interval with probability 0.95, until I pointed out that $\mu$ is just a point value and doesn't have a probability distribution associated with it. Morey and Wagenmakers and Rouder et al have been shouting themselves hoarse about confidence intervals, and how many people don't understand them, also see this paper. Ironically, psychologists have responded to these complaints through various media, but even this response only showcases how psychologists have only a partial and misconstrued understanding of confidence intervals. I feel that part of the problem is that scientists hate to back off from a position they have taken, and so they tend to hunker down and defend defend defend their position. From the perspective of a statistician who understands both Bayes and frequentist positions, the conclusion would have to be that Morey et al are right, but for large sample sizes, the difference between a credible interval and a confidence interval (I mean the actual values that you get for the lower and upper bound) are negligible. You can see examples in our recently ArXiv'd paper.
Third, I tried to explain that there is a cultural difference between statisticians on the one hand and (most) psychologists and almost all psychologist, linguists, etc. on the other. For the latter group (with the obvious exception of people using Bayesian methods for data analysis), the whole point of fitting a statistical model is to do a hypothesis test, i.e., to get a p-value out of it. They simply do not care what the assumptions and internal moving parts of a t-test or a linear mixed model are. I know lots of users of lmer who are focused on one and only one thing: is my effect significant? I have repeatedly seen experienced experimenters in linguistics simply ignoring the independence assumption of data points when doing a paired t-test; people often do paired t-tests on unaggregated data, with multiple rows of data points for each subject (for example). This leads to spurious significance effects, which they happily and unquestioningly accept because that was the whole goal of the exercise. I show some examples in my lecture2 slides (slide 70).
It's not just linguists, you can see the consequences of ignoring the independence assumption in this reanalysis of the infamous study on how future tense marking in language supposedly influences economic decisions. Once the dependencies between languages are taken into account, the conclusion that Chen originally drew doesn't really hold up much:
" When applying the strictest tests for relatedness, and when data is not aggregated across individuals, the correlation is not significant."
Similarly, Amy Cuddy et al's study on how power posing increases testosterone levels also got published only because the p value just scraped in below 0.05 at 0.045 or so. You can see in their figure 3 reporting the testosterone increase
that their confidence intervals are huge (this is probably why they report standard errors, it wouldn't look so impressive if they had reported CIs). All they needed to show to make their point was to get the p-value below 0.05. The practical relevance of a 12 picogram/ml increase in testosterone is left unaddressed. Another recent example from Psychological Science, which seems to publish studies that might attract attention in the popular press, is this study on how ovulating women wear red. This study is a follow up on the notorious Psychological Science study by Beall and Tracy. In my opinion, the Beall and Tracy study reports a bogus result because they claim that women wear red or pink when ovulating, but when I reanalyzed their data I found that the effect was driven by pink alone. Here is my GLM fit for red or pink, red only and pink only. You can see that the "statistically significant" effect is driven entirely by pink, making the title of their paper (Women Are More Likely to Wear Red or Pink at Peak Fertility) true only if you allow the exclusive-or reading of the disjunction:
The new study by Eisenbruch et al reports a statistically significant effect on this red-pink issue, but now it's only about red:
"A mixed regression model confirmed that, within subjects, the odds of wearing red were higher during the estimated fertile window than on other cycle days, b = 0.93, p = .040, odds ratio (OR) = 2.53, 95% confidence interval (CI) = [1.04, 6.14]. The 2.53 odds ratio indicates that the odds of wearing a red top were about 2.5 times higher inside the fertile window, but there was a wide confidence interval."
To their credit, they note that their confidence interval is huge, and essentially includes 1. But since the p-value is below 0.05 this result is considered evidence for the "red hypothesis". It may well be that women who are ovulating wear red; I have no idea and have no stake in the issue. Certainly, I am not about to start looking at women wearing red as potential sexual partners (quite independent from the fact that my wife would probably kill me if I did). But it would be nice if people would try to do high powered studies, and report a replication in the same study they publish. Luckily nobody will die if these studies report mistaken results, but the same mistakes are happening in medicine, where people will die as a result of incorrect conclusions being drawn.
All these examples show why the focus on p-values is so damaging for answering research questions.
Not surprisingly, for the statistician, the main point of fitting a model (even in a confirmatory factorial analysis) is not to derive a p-value from it; in fact, for many statisticians the p-value may not even rise to consciousness. The main point of fitting a model is to define a process which describes, in the most economical way possible, how the data were generated. If the data don't allow you to estimate some of the parameters, then, for a statistician it is completely reasonable to back off to defining a simpler generative process.
This is what Gelman and Hill also explain in their 2007 book (italics mine). Note that they are talking about fitting Bayesian linear mixed models (in which parameters like correlations can be backed off to 0 by using appropriate priors; see the Stan code using lkj priors here), not frequentist models like lmer. Also, Gelman would never, ever compute a p-value.
Gelman and Hill 2007, p. 549:
For the statistician, simplicity of expression and understandability of the model (in the Gelman and Hill sense of being able to derive sensible posterior (predictive) distributions) are of paramount importance. For the psychologist and linguist (and other areas), what matters is whether the result is statistically significant. The more vigorously you can reject the null, the more excited you get, and the language provided for this ("highly significant") also gives the illusion that we have found out something important (=significant).
This seems to be a fundamental disconnect between statisticians, and end-users who just want their p-value. A further source of the disconnect is that linguists and psychologists etc. look for cookbook methods, what a statistician I know once derisively called a "one and done" approach. This leads to blind data fitting: load data, run single line of code, publish result. No question ever arises about whether the model even makes sense. In a way this is understandable; it would be great if there was a one-shot solution to fitting, e.g., linear mixed models. It would simplify life so much, and one wouldn't need to spend years studying statistics before one can do science. However, the same scientists who balk at studying statistics will willingly spend time studying their field of expertise. No mainstream (by which I mean Chomskyan) syntactician is going to ever use commercial software to print out his syntactic derivation without knowing anything about the syntactic theory. Yet this is exactly what these same people expect from statistical software, to get an answer without having any understanding of the underlying statistical machinery.
The bottom line that I tried to convey in my course was: forget about the p-value (except to soothe the reviewer and editor and to build your career), focus on doing high powered studies, check model assumptions, fit appropriate models, replicate your findings, and publish against your own pet theories. Understanding what all these words means requires some study, and we should not shy away from making that effort.
PS I am open to being corrected---like everyone else, I am prone to making mistakes. Please post corrections, but with evidence, in the comments section. I moderate the comments because some people post spam there, but I will allow all non-spam comments.
PPS The teaching evaluation for this course just came in from ESSLLI; here it is. I believe 5.0 is a perfect score.
All materials for the first week are available on github, see here.
The frequentist course went well, but the Bayesian course was a bit unsatisfactory; perhaps my greater experience in teaching the frequentist stuff played a role (I have only taught Bayes for three years). I've been writing and rewriting my slides and notes for frequentist methods since 2002, and it is only now that I can present the basic ideas in five 90 minute lectures; with Bayes, the presentation is more involved and I need to plan more carefully, interspersing on-the-spot exercises to solidify ideas. I will comment on the Bayesian Data Analysis course in a subsequent post.
The first week (five 90 minute lectures) covered the basic concepts in frequentist methods. The audience was amazing; I wish I always had students like these in my classes. They were attentive, and anticipated each subsequent development. This was the typical ESSLLI crowd, and this is why teaching at ESSLLI is so satisfying. There were also several senior scientists in the class, so hopefully they will go back and correct the misunderstandings among their students about what all this Null Hypothesis Significance Testing stuff gives you (short answer: it answers *a* question very well, but it's the wrong question, nothing that is relevant to your research question).
I won't try to summarize my course, because the web page is online and you can also do exercises on datacamp to check your understanding of statistics (see here). You get immediate feedback on your attempts.
Stepping away from the technical details, I tried to make three broad points:
First, I spent a lot of time trying to clarify what a p-value is and isn't, focusing particularly on the issue of Type S and Type M errors, which Gelman and Carlin have discussed in their excellent paper.
Here is the way that I visualized the problems of Type S and Type M errors:
What we see here is repeated samples from a Normal distribution with true mean 15 and a typical standard deviation seen in psycholinguistic studies (see slide 42 of my slides for lecture2). The horizontal red line marks the 20% power line; most psycholinguistic studies fall below that line in terms of power. The dramatic consequence of this low power is the hugely exaggerated effects (which tend to get published in major journals because they also have low p-values) and the remarkable proportion of cases where the sample mean is on the wrong side of the true value 15. So, you are roughly equally likely to get a significant effect with a sample mean smaller to much smaller than the true mean, or larger and much larger than the true mean. With low power, regardless of whether you get a significant result or not, if power is low, and it is in most studies I see in journals, you are just farting in a puddle.
It is worth repeating this: once one considers Type S and Type M errors, even statistically significant results become irrelevant, if power is low. It seems like these ideas are forever going to be beyond the comprehension of researchers in linguistics and psychology, who are trained to make binary decisions based on p-values, weirdly accepting the null if p is greater than 0.05 and, just as weirdly, accepting their favored alternative if p is less than 0.05. The p-value is a truly interesting animal; it seems that a recent survey of some 400 Spanish psychologists found that, despite their being active in the field for quite a few years on average, they had close to zero understanding of what a p-value gives you. Editors of top journals in psychology routinely favor lower p-values, because they mistakenly think this makes "the result" more convincing; "the result" is the favored alternative. So even seasoned psychologists (and I won't even get started with linguists, because we are much, much worse), with decades of experience behind them, often have no idea what the p-value actually tells you.
A remarkable misunderstanding regarding p-values is the claim that it tells you whether the effect was "by chance". Here is an example from Replication Index's blog:
"The Test of Insufficient Variance (TIVA) shows that the variance in z-scores is less than 1, but the probability of this event to occur by chance is 10%, Var(z) = .63, Chi-square (df = 11) = 17.43, p = .096."
Here is another example from a self-published manuscript by Daniel Ezra Johnson:
"If we perform a likelihood-ratio test, comparing the model with gender to a null model with no predictors, we get a p-value of 0.0035. This implies that it is very unlikely that the observed gender difference is due to chance."
One might think that the above examples are not peer-reviewed, and that peer review would catch such mistakes. But even people explaining p-values in publications are unable to understand that this is completely false. An example is Keith Johnson's textbook, Quantitative Methods in Linguistics, which repeatedly talks about "reliable effects" and effects which are and are not due to chance. It is no wonder that the poor psychologist/linguist thinks, ok, if the p-value is telling me the probability that the effect is due to chance, and if the p-value is low, then the effect is not due to chance and the effect must be true. The mistake here is that the p-value is telling you the probability of the result being "due to chance" conditional on the null hypothesis being true. It's better to explain the p-value as the probability of getting the statistic (e.g., t-value) or something more extreme, under the assumption that the null hypothesis is true. People seem to drop the italicized part and this starts to propagate the misunderstanding for future generations. To repeat, the p-value is a conditional probability, but most people interpret it as an unconditional probability because they drop the phrase "under the null hypothesis" and truncate the statement to be about effects being due to chance.
Another bizarre thing I have repeatedly seen is misinterpreting the p-value as Type I error. Type I error is fixed at a particular value (0.05) before you run the experiment, and is the probability of your incorrectly rejecting the null when it's true, under repeated sampling. The p-value is what you get from your single experiment and is the conditional probability of your getting the statistic you got or something more extreme, assuming that the null is true. Even this point is beyond comprehension for psychologists (and of course linguists). Here is a bunch of psychologists explaining in an article why a p=0.000 should not be reported as an exact value:
"p = 0.000. Even though this statistical expression,
used in over 97,000 manuscripts according to Google Scholar,
makes regular cameo appearances in our computer printouts, we
should assiduously avoid inserting it in our Results sections. This
expression implies erroneously that there is a zero probability
that the investigators have committed a Type I error, that is, a
false rejection of a true null hypothesis (Streiner, 2007). That
conclusion is logically absurd, because unless one has examined
essentially the entire population, there is always some chance
of a Type I error, no matter how meager. Needless to say, the
expression “p < 0.000” is even worse, as the probability of
committing a Type I error cannot be less than zero. Authors
whose computer printouts yield significance levels of p = 0.000
should instead express these levels out to a large number of
decimal places, or at least indicate that the probability level is
below a given value, such as p < 0.01 or p < 0.001."
The p-value is the probability of committing a Type I error, eh? It is truly embarrassing that people who are teaching this stuff have distorted the meaning of the p-value so drastically and just keep propagating the error. I should mention though that this paper I am citing appeared in Frontiers, which I am beginning to question as a worthwhile publication venue. Who did the peer review on this paper and why did they not catch this basic mistake?
Even Fisher (p. 16 of The Design of Experiments, Second Edition, 1937) didn't buy the p-value; he is advocating for replicability as the real decisive tool:
"It is usual and convenient for experimenters to take-5 per cent. as a standard level of significance, in the sense that they are prepared to ignore all results which fail to reach this standard, and, by this means, to eliminate from further discussion the greater part of the fluctuations which chance causes have introduced into their experimental results. No such selection can eliminate the whole of the possible effects of chance. coincidence, and if we accept this convenient convention, and agree that an event which would occur by chance only once in 70 trials is decidedly" significant," in the statistical sense, we thereby admit that no isolated experiment, however significant in itself, can suffice for the experimental demonstration of any natural phenomenon; for the "one chance in a million" will undoubtedly occur, with no less and no more than its appropriate frequency, however surprised we may be that it should occur to us. In order to assert that a natural phenomenon is experimentally demonstrable we need, not an isolated record, but a reliable method of procedure. In relation to the test of significance, we may say that a phenomenon is experimentally demonstrable when we know how to conduct an experiment which will rarely fail to give us a statistically significant result."
Second, I tried to clarify what a 95% confidence interval is and isn't. At least a couple of students had a hard time accepting that the 95% CI refers to the procedure and not that the true $\mu$ lies within one specific interval with probability 0.95, until I pointed out that $\mu$ is just a point value and doesn't have a probability distribution associated with it. Morey and Wagenmakers and Rouder et al have been shouting themselves hoarse about confidence intervals, and how many people don't understand them, also see this paper. Ironically, psychologists have responded to these complaints through various media, but even this response only showcases how psychologists have only a partial and misconstrued understanding of confidence intervals. I feel that part of the problem is that scientists hate to back off from a position they have taken, and so they tend to hunker down and defend defend defend their position. From the perspective of a statistician who understands both Bayes and frequentist positions, the conclusion would have to be that Morey et al are right, but for large sample sizes, the difference between a credible interval and a confidence interval (I mean the actual values that you get for the lower and upper bound) are negligible. You can see examples in our recently ArXiv'd paper.
Third, I tried to explain that there is a cultural difference between statisticians on the one hand and (most) psychologists and almost all psychologist, linguists, etc. on the other. For the latter group (with the obvious exception of people using Bayesian methods for data analysis), the whole point of fitting a statistical model is to do a hypothesis test, i.e., to get a p-value out of it. They simply do not care what the assumptions and internal moving parts of a t-test or a linear mixed model are. I know lots of users of lmer who are focused on one and only one thing: is my effect significant? I have repeatedly seen experienced experimenters in linguistics simply ignoring the independence assumption of data points when doing a paired t-test; people often do paired t-tests on unaggregated data, with multiple rows of data points for each subject (for example). This leads to spurious significance effects, which they happily and unquestioningly accept because that was the whole goal of the exercise. I show some examples in my lecture2 slides (slide 70).
It's not just linguists, you can see the consequences of ignoring the independence assumption in this reanalysis of the infamous study on how future tense marking in language supposedly influences economic decisions. Once the dependencies between languages are taken into account, the conclusion that Chen originally drew doesn't really hold up much:
" When applying the strictest tests for relatedness, and when data is not aggregated across individuals, the correlation is not significant."
Similarly, Amy Cuddy et al's study on how power posing increases testosterone levels also got published only because the p value just scraped in below 0.05 at 0.045 or so. You can see in their figure 3 reporting the testosterone increase
that their confidence intervals are huge (this is probably why they report standard errors, it wouldn't look so impressive if they had reported CIs). All they needed to show to make their point was to get the p-value below 0.05. The practical relevance of a 12 picogram/ml increase in testosterone is left unaddressed. Another recent example from Psychological Science, which seems to publish studies that might attract attention in the popular press, is this study on how ovulating women wear red. This study is a follow up on the notorious Psychological Science study by Beall and Tracy. In my opinion, the Beall and Tracy study reports a bogus result because they claim that women wear red or pink when ovulating, but when I reanalyzed their data I found that the effect was driven by pink alone. Here is my GLM fit for red or pink, red only and pink only. You can see that the "statistically significant" effect is driven entirely by pink, making the title of their paper (Women Are More Likely to Wear Red or Pink at Peak Fertility) true only if you allow the exclusive-or reading of the disjunction:
The new study by Eisenbruch et al reports a statistically significant effect on this red-pink issue, but now it's only about red:
"A mixed regression model confirmed that, within subjects, the odds of wearing red were higher during the estimated fertile window than on other cycle days, b = 0.93, p = .040, odds ratio (OR) = 2.53, 95% confidence interval (CI) = [1.04, 6.14]. The 2.53 odds ratio indicates that the odds of wearing a red top were about 2.5 times higher inside the fertile window, but there was a wide confidence interval."
To their credit, they note that their confidence interval is huge, and essentially includes 1. But since the p-value is below 0.05 this result is considered evidence for the "red hypothesis". It may well be that women who are ovulating wear red; I have no idea and have no stake in the issue. Certainly, I am not about to start looking at women wearing red as potential sexual partners (quite independent from the fact that my wife would probably kill me if I did). But it would be nice if people would try to do high powered studies, and report a replication in the same study they publish. Luckily nobody will die if these studies report mistaken results, but the same mistakes are happening in medicine, where people will die as a result of incorrect conclusions being drawn.
All these examples show why the focus on p-values is so damaging for answering research questions.
Not surprisingly, for the statistician, the main point of fitting a model (even in a confirmatory factorial analysis) is not to derive a p-value from it; in fact, for many statisticians the p-value may not even rise to consciousness. The main point of fitting a model is to define a process which describes, in the most economical way possible, how the data were generated. If the data don't allow you to estimate some of the parameters, then, for a statistician it is completely reasonable to back off to defining a simpler generative process.
This is what Gelman and Hill also explain in their 2007 book (italics mine). Note that they are talking about fitting Bayesian linear mixed models (in which parameters like correlations can be backed off to 0 by using appropriate priors; see the Stan code using lkj priors here), not frequentist models like lmer. Also, Gelman would never, ever compute a p-value.
Gelman and Hill 2007, p. 549:
"Don’t get hung up on whether a coefficient “should” vary by group. Just allow it
to vary in the model, and then, if the estimated scale of variation is small (as with
the varying slopes for the radon model in Section 13.1), maybe you can ignore it if
that would be more convenient.
Practical concerns sometimes limit the feasible complexity of a model—for example, we might fit a varying-intercept model first, then allow slopes to vary, then add group-level predictors, and so forth. Generally, however, it is only the difficulties of fitting and, especially, understanding the models that keeps us from adding even more complexity, more varying coefficients, and more interactions."
Practical concerns sometimes limit the feasible complexity of a model—for example, we might fit a varying-intercept model first, then allow slopes to vary, then add group-level predictors, and so forth. Generally, however, it is only the difficulties of fitting and, especially, understanding the models that keeps us from adding even more complexity, more varying coefficients, and more interactions."
For the statistician, simplicity of expression and understandability of the model (in the Gelman and Hill sense of being able to derive sensible posterior (predictive) distributions) are of paramount importance. For the psychologist and linguist (and other areas), what matters is whether the result is statistically significant. The more vigorously you can reject the null, the more excited you get, and the language provided for this ("highly significant") also gives the illusion that we have found out something important (=significant).
This seems to be a fundamental disconnect between statisticians, and end-users who just want their p-value. A further source of the disconnect is that linguists and psychologists etc. look for cookbook methods, what a statistician I know once derisively called a "one and done" approach. This leads to blind data fitting: load data, run single line of code, publish result. No question ever arises about whether the model even makes sense. In a way this is understandable; it would be great if there was a one-shot solution to fitting, e.g., linear mixed models. It would simplify life so much, and one wouldn't need to spend years studying statistics before one can do science. However, the same scientists who balk at studying statistics will willingly spend time studying their field of expertise. No mainstream (by which I mean Chomskyan) syntactician is going to ever use commercial software to print out his syntactic derivation without knowing anything about the syntactic theory. Yet this is exactly what these same people expect from statistical software, to get an answer without having any understanding of the underlying statistical machinery.
The bottom line that I tried to convey in my course was: forget about the p-value (except to soothe the reviewer and editor and to build your career), focus on doing high powered studies, check model assumptions, fit appropriate models, replicate your findings, and publish against your own pet theories. Understanding what all these words means requires some study, and we should not shy away from making that effort.
PS I am open to being corrected---like everyone else, I am prone to making mistakes. Please post corrections, but with evidence, in the comments section. I moderate the comments because some people post spam there, but I will allow all non-spam comments.
PPS The teaching evaluation for this course just came in from ESSLLI; here it is. I believe 5.0 is a perfect score.
Statistical methods for linguistic research: Foundational Ideas (Vasishth)
| Lecturer1 | 4.9 |
| Did the course content correspond to what was proposed? | 4.9 |
| Course notes | 4.6 |
| Session attendance | 4.4 |
(19 respondents)
- Very good course.
- The lecturer was simply great. He has made hard concepts really easy to understand. He also has been able to keep the class interested. A real pity to miss the last lecture !!
- The only reason that this wasn't the best statistics course is that I had a great lecturer at my university on this. Very entertaining, informative, and correct lecture, I can't think of anything the lecturer could do better.
- Informative, deep and witty. Simply awesome.
- Professor Shravan Vasishth was hands down the best lecturer at ESSLLI 2015. I envy the people who actually get to learn from him for a whole semester instead of just a week or two. The course was challenging for someone with not much background in statistics, but Professor Vasishth provided a bunch of additional material. He's the best!
- Great course, very detailed explanations and many visual examples of some statistical phenomena. However, it would be better to include more information on regression models, especially with effects (model quality evaluation, etc) and more examples of researches from linguistic field.
- It was an extremely useful course presented by one of the best lecturers I've ever met. Thank you!
- Amazing course. Who would have thought that statistics could be so interesting and engaging? Kudos to lecturer Shravan Vasishth who managed to condense so much information into only 5 sessions, who managed to filter out only the most relevant things that will be applicable and indeed used by everyone who attendet the course and who managed to show the usefulness of the material. A great lecturer who never went on until everything was cleared up and made even the most daunting of statistical concepts seem surmountable. The only thing I'm sorry for is not having the opportunity to take his regular, semester-long statistics course so I can enjoy a more in depth look at the material and let everything settle properly. Five stars, would take again.
- Absolutely great!!
Saturday, February 14, 2015
Getting a statistics education: Review of the MSc in Statistics (Sheffield)
 [This post was written between Sept 2012 and Feb 2015. I will post an update in Sept. 2015]
[This post was written between Sept 2012 and Feb 2015. I will post an update in Sept. 2015] Last edit: June 27, 2015
Final edit: Nov 3, 2015 (added MSc thesis grade)
Some background:
I started using statistics for my research sometime in 1999 or 2000. I was a student at Ohio State, Linguistics, and I had just gotten interested in psycholinguistics. I knew almost nothing about statistics at that time. I did one Intro to Stats course in my department with Mike Broe (4 weeks), and that was it. In 1999 I developed repetitive strain injury, partly from using Excel and SPSS, and started googling for better statistical software. Someone pointed me to |stat, but eventually I found R. That was a transformative moment.
The next stage in my education came in 2000, when I decided to go to the Statistical Consulting department at OSU and showed them my repeated measure ANOVA analyses. The response I got was: why are you fitting ANOVAs? You need linear mixed models. The statisticians showed me what I had to do code-wise, and I went ahead and finished my dissertation work using the nlme package. The Pinheiro and Bates book had just come out then and I got myself a copy, understanding almost nothing in the book beyond the first few chapters.
After that, I published a few more papers on sentence processing using nlme and then lmer, and in 2011 I co-wrote a book with Mike Broe (the basic template of the book was based on his lecture notes at OSU, he had used Mathematica or something like that, but I used R and expanded on his excellent simulation-based approach). This book revealed the incompleteness of my understanding, as spelled out in the scathing (and well-deserved) critique by Christian Robert. Even before this review came out, I had already realized in early 2011 that I didn't really understand what I was doing. My sabbatical was coming up in winter 2011, and I enrolled for the graduate certificate in statistics at Sheffield to get a better understanding of statistical theory. Here is my review of the distance-based graduate certificate in statistics taught at Sheffield.
At the end of that graduate certificate, I felt that I still didn't really understand much that was of practical relevance to my life as a researcher. That led me to do the MSc in Statistics at Sheffield, which I have been doing over three years (2012-15). This is a review of the MSc program.
Short version of this review: The three year distance MSc program at Sheffield is outstanding. I highly recommend it to anyone wanting to acquire a good, basic understanding of statistical theory and inference. You can alternatively do the course over two years (probably impossible or very hard if you are also working full time, like me), or over one year full time (I don't know how people can do the degree in one year and still enjoy it). Be prepared to work hard and to find your own answers.
Long version:
Cost: For EU citizens, the three-year part time program costs about 2000 British pounds a year, not including the travel costs to get to Sheffield for the annual exams and presentations. For non-EU citizens, it's about 5000 pounds a year, still cheaper than most US programs.
Summary notes of the MSc program: I made summary notes for the exams during the three years. These are still very much in progress and are available from:
https://github.com/vasishth/MScStatisticsNotes
The courses I found most interesting and practically useful for my own research were Linear Modelling, Inference (Bayesian Statistics and Computational Inference), Medical Statistics, and Dependent data (Multivariate Analysis).
Course structure: Over three years, one does two courses each year, plus a dissertation. One has to commit about 15-20 hours a week in the 3-year program, although I think I did not do that much work, more like 12 hours a week on average (I had a lot of other work to do and just didn't have enough time to devote to statistics). There are four 3 hour sort-of open book exams that one has to go to Sheffield for, plus a group oral presentation, a simulated consultation, and project submissions. Every course has regular assignments/projects, all are graded but only a subset count for the final exam (15% of the final grade). The minimum you have to get to pass is 50%.
The MSc program is taught to residential students and to distance students in parallel: the residentials are there in Sheffield, attending lectures etc. The distance students follow the course over a mailing list. So, someone like me, who's doing the course over three years, is going to overlap with three batches of the MSc residential students. This has the effect that one has no classmates one knows, except maybe others who are doing the same three-year sequence with you.
The exams, which are the most stressful part of the program, are open book in that one can bring lecture notes and one's own but no textbooks. However, the exams are designed in such a way that if you don't already know the material inside out, there is almost no point in taking lecture notes in with you---there won't be enough time to look up the notes. I did take the official lecture notes with me for the first three exams, but I never once opened them. Instead, I only relied on my own summary sheets. Also, the exams are designed so that most people can't finish the required questions (any 5 out of 6) in the three hours. At least I never managed to finish all the questions to my satisfaction in any exam.
The first year (2012-13)
The first year courses were 6002 (Stats Lab) and 6003 (Linear Modelling). There was a project-based assessment for the first, and a 3 hour exam for the second.
6002 (Stats Lab): most of the course was about learning R, which anyone who had done the grad certificate did not need. It was only in the last weeks that things got interesting, with optimization. I didn't like the notes on optimization and MLE much, though. There wasn't enough detail, and I had to go searching in books and on the internet to find comprehensive discussions. Here I would recommend Ben Bolker's chapters 6-8, which are on his web page, complete with .Rnw files. Also, I just found a neat looking book (not read yet) which I wish I had had in 2012: Modern Optimization with R.
Overall the Stats Lab course had the feel of an intro to R, which is what it should have been called. It should have been possible to test out of such a course---I did not need to read the first 12 of 13 chapters over 9 months, I could have done it in a week or less, I'm sure that's true for those of my classmates who did the graduate certificate. However, I do see the point of the course for non-R users. I guess this is the perennial problem of teaching; students come in with different levels, you have to cater to the lowest common denominator. Also, the introduction to R is pretty dated and needs a major overhaul. Much has happened since Hadley Wickham arrived on the scene, and it's a shame not to use his packages. Finally, the absence of literate programming tools was surprising to me. I expected it to be a standard operating procedure in statistics to use Sweave or the like.
6003 (Linear Modelling): this course was absolutely amazing. The lecture notes were very well-written and very detailed (with some exceptions, noted below). Linear mixed models didn't get a particularly detailed treatment; I would have preferred a matrix presentation of LMM theory, and would have liked to learn how to implement these models myself.
Some problems I faced in year 1:
One issue in the course was the slow return of corrected assignments. By the time the assignment comes back graded (well, we just get general feedback and a grade), you've forgotten the details. Another strange aspect is that the grades for assignments were sometimes sent by regular air-mail. This was surprising in an online course.
One frustrating aspect of the courses was that a number of statements were made without any justification, proof, or further explanation. Example: "In R the default choice is the corner-point constraints given above, but in SPlus the default is the Helmert form, which is more convenient computationally, though more difficult to interpret." Wow, I want to know more! But this point is never discussed again. One consequence is a feeling that one must simply take certain facts as given (or work it out yourself). I think it would have been helpful to point the interested student to a reference.
The responses to questions on the mailing list are sometimes slow to come. Answers to questions asked online sometimes didn't really address the question, and one was left in the same state of uncertainty as earlier (a familiar feeling when you talk to a statistician!).
Where the graduate certificate shone was in the excruciatingly detailed feedback; this was where I learnt the most in that course. By contrast, the feedback to some of the assignments was pretty sketchy. I never really knew what a perfect solution would have looked like.
Of course, I can see why all this happens: professors are busy, and not always able to respond quickly to questions. I myself am sometimes just as slow to respond as a teacher; I guess I need to work on that aspect of my own teaching.
My final marks in these first-year courses were 63 per cent in each course.
The second year (2013-14)
The second year courses were 6001 (Data Analysis) and 6004 (Inference: Bayesian Statistics and Computational Inference). There was a project-based assessment for the first, and a 3 hour exam for the second.
In Data Analysis we did several projects which simulated real-life consulting, or involved doing actual experiments (e.g., building aeroplanes). There was one project where one had to choose a news media article about a piece of scientific work, and then compare it with the actual scientific work. The consulting project didn't work so well for me, because we were teamed up in fives and we didn't know each other. It was very hard to coordinate a project when all your colleagues are unknown to you, and email is the only way to communicate.
For the news media article, I chose the article Gelman attacked on his blog, about women wearing red to signal sexual availability. It was interesting because the claims in the Psych Science didn't really pan out. I reanalyzed the original data, and found that the effect was driven by pink, not red; the authors had recoded red and pink as red or pink, presumably in order to make the claim that women wear reddish hues. It's hard to believe that this was not a post-hoc step after seeing the data (although I think the authors claim it was not---I suppose it's possible that it wasn't); after all, if they had originally intended to treat red and pink as one unit color type, then why did they have two columns, one for red and one for pink?
The Data Analysis course was definitely not challenging; it was rather below the level of data analysis I have to do in my own research. However, I was thankful not to be overloaded in this course because the Bayesian analysis course took up all my energy in my second year.
The course on Bayesian statistics was a whole other animal. I read a lot of books that were not assigned as required readings (mostly, Gelman et al's BDA3, and Lunn et al, but also Lynch's excellent textbook). I did all the three exercises that were assigned (these are graded but do not count for the final grade). My scores were 20/20, 22/30, 23/30. I never really understood what exactly led to those points being lost; not much detailed explanation was provided. One doesn't know how many marks one loses for making a figure too small, for example (I was following Gelman's example of showing lots of figures, which requires making them smaller, but evidently this was frowned upon). As is typical for this degree program, the grading is pretty harsh and tight-lipped (the harsh grading is not a bad thing; but the lack of information on what to improve in the answer was frustrating).
The Bayesian lecture notes could be improved. They seem to have a disjointed feel; perhaps they were written by different people. The Bayesian lecture notes were very different than, say, the linear modeling notes, which really drilled the student on practical details of model fitting. In the Bayesian course, there were sudden transitions to topics that fizzled out quickly and were never resurrected. An example is decision theory; one section starts out defining some basic concepts, and then quickly ends. Inference and decision theory was never discussed. There were sections that were in the notes but not needed for the exams; for an MSc level program I would have wanted to read that material (and did). I had some questions on these non-examinable sections, but never could get an answer, which was pretty frustrating.
The biggest thing that could be improved in these lecture notes is to provide more contact with code. Unfortunately, WinBUGS was introduced, and very late in the course, and then a fairly major project (which counts for the final grade) was assigned that was based entirely on modeling in WinBUGS. Apart from the fact that WinBUGS is just not a well-designed software (JAGS or Stan is much better), not much practice was given in fitting models, certainly not as much as was given for linear modelling. Model fitting should be an integral part of the course from the outset, and WinBUGS should be abandoned in favor of JAGS.
If I had not done a lot of reading on my own, and not learnt JAGS and Stan, I would have really suffered in this course. Maybe that's what the lecture notes are intending to do: it's a graduate-level course, and maybe the expectation is that one looks up the details on one's own.
As it was, I enjoyed doing the Bayesian exercises, which were very neat problems---just hard enough to make you think, but not so hard that you can't solve them if you think hard and do your own research.
One thing that was never discussed in the Bayesian data analysis course was how to do statistical inference, for example in factorial $2\times 2$ repeated measures designs. Textbooks on Bayesian methods don't discuss this either; perhaps they consider it enough that you get the posterior; you can draw your own conclusions from that.
I got scores in the mid 60s for each course. I think I had 63 in Data Analysis and 67 in Inference.
The third year
The third year courses were MAS6011 (Dependent data) and MAS6012 (Sampling, Design, Medical Statistics). There is a 3 hour exam for each course.
The dependent data course was truly amazing. In the first semester, I got to grips with multivariate analysis, and with some interesting data mining type of tools such as PCA and linear discriminant analysis. The lecture notes could have been a lot more detailed for a graduate program; the lack of detail was probably due to the fact that undergrads and grad students were mixed in in the same class. The second semester was about time-series analysis, and was the best taught course and the most exciting I took in this MSc. For the first time, video lectures are being provided every week, and these are proving to be extremely helpful.
What really resonated with me in this course was state space modeling. I wish the whole course had been about that topic; the ARIMA modeling framework of Box and Jenkins is really amazing but pales in significance when you see what SSMs can do. Maybe it would have been better to teach a two-semester sequence instead of compression a Data Mining type of course into the first semester, and TS into the second. I would happily have done another course instead of doing those Stats Labs and such like "soft" courses, as I mention elsewhere.
The Medical Statistics course was fascinating because it was here that one finally saw issues being dealt with where people's lives would be at stake depending on the answer we obtain. One amazing fact I discovered is that Pocock 1983 considers power below 70% in an experiment to be unethical. Psycholinguists and psychologists routinely run low power studies and publish their null results in prestigious journals. Luckily nobody will die as a result of these studies! Another amazing fact is that frequentist statistics is standard practice in medicine. I would have expected that Bayesian stats would dominate in such a vitally important application of statistics. I am willing to use p-values to make a binary decision to help a journal editor feel good about the paper, but not if I am deciding whether drug X will help stave off death for a patient. I am really glad that I do not need to enter the job market as a statistician. If I were starting out my career after finishing this degree, I would probably have done into a pharma company, and it is horrifying to think that I would be forced to deliver p-values as decision-making tool.
For the first semester, the medstats lecture notes were not that well written, with not much detail, full of typos and bullet point type presentations. The slides had no page numbers. These lecture notes and slides need a major overhaul in my opinion. I didn't get any detailed feedback on the first two exercises I submitted, and the feedback I did get I could not read as it was handwritten with one of those ball-point pens that don't steadily deliver ink. The feedback, such as it was, came in unusually late as well. By contrast, the survival analysis lecture notes were much better, and I learnt a lot.
The second semester lecture notes and slides were on the design of experiments and sampling theory (stratified sampling, cluster sampling, capture-recapture sampling, etc.). The DoE part was outstanding; for the first time, I learnt how optimal experimental design is set out, and learnt to determine optimality of design using the General Equivalence Theorem. I think I would have liked to have this course right after Linear Modelling (in the three year distance program, this course and LM are separate by a year of coursework on computational statistics and Bayesian Data Analysis), although the gap did have one advantage that linear modeling theory had some time to sink in before I studied experiment design. I was less excited by the sampling part, but I think that this is because I am probably never going to be doing sample surveys. I just couldn't whip up enough enthusiasm for that topic, but I did hunker down and learn everything anyway. The second semester also came with weekly video recordings, so for the first time I was able to watch the same lecture that the residential students were attending.
Update:
I got 67% in Medical Statistics and 70% in Dependent Data (a distinction, my first in this MSc program!).
This was much better than I expected; I write slowly (I enjoy writing with my high quality gold plated, lacquered Namiki Pilot fountain pen, and the sheer pleasure of having the pen glide over paper, leaving mesmerizing, exquisite strokes of black ink, slows me down a lot), and so I knew I would not be able to finish the papers, and I didn't. But I guess the questions that I answered I must have done reasonably OK. For these two exams, I practiced a lot more with the hand calculator too, and I noticed that practice makes me... well, not perfect, but better. I did stop making stupid mistakes like forgetting that log is by default to the base 10 and I have explicitly ask for a log_e, and mistyping multiplication when I meant division. (In the BDA exam I actually managed to get a probability greater than 1 in one answer due to this kind of idiocy. Since I didn't have time to go back to fix my mistake, I just wrote "doesn't make sense, there must be a calculation error somewhere", hoping that the grader will realize that I understand the method but can't type on a hand-held).
Final Update on the MSc Dissertation:
There's also a thesis to be written as part of the MSc; that counts for 60 credits in the 180 credit MSc program. I would have preferred to do more coursework than do the thesis, but I can see why a thesis is required (all our programs in Potsdam require them too). More on that in September or October 2015.
The thesis work went quite well overall. Initially I ran into a very difficult situation that turned out to be due to my having coded up my model incorrectly. My advisor (Jeremy Oakley) was extremely responsive and deftly asked me the right questions that led me to find the bugs in my code; after that it was smooth sailing. That experience really showed (if that's not obvious) that it helps to work with an expert in the field. My final grade on the thesis was 73%.
General comments/suggestions for improvement:
1. The MSc currently has three specializations: Statistics, Medical Statistics, and Financial Statistics. Each has slightly different requirements (e.g., for Financial, you need to demonstrate specific math ability). I would add a fourth specialization, to reflect the needs of statisticians today. This could be called Computational Statistics or something like that.
In this specialization, one could require a background in R programming, just as Financial Stats requires advanced math. One could replace Stats lab and Data Analysis with a course on Statistical Computing (following some subset of the contents of textbooks like Eubank et al, Eddenbeutel, Cortez, Hadley), and Statistical Learning (aka Data Mining), following a textbook like James et al. I am sure that such a specialization is badly needed; see, for example, the puzzled question asked by a statistician not so long ago in AMSTAT news: Aren't we data science? One can't prepare statisticians as "data scientists" if they don't have serious computing ability.
Some of the data mining related materials turns up in Dependent Data in year 3, and that's fine; there is much more that one needs exposure to today. For me, the Stats Lab and Data Analysis courses did not have enough bang for the buck. I can see that such courses could be useful to newcomers to R and data analysis (but at the grad level, I find it hard to believe that the student would have never seen R; I guess it's possible).
But these courses didn't really challenge me to deal with real-life problems one might be likely to encounter as a future statistician (writing one's own packages, solving large-scale data mining problems). If there had been a more computationally oriented stream which assumed R, I would have taken that route.
Some MS(c) programs with the kind of focus I am suggesting:
a. St Andrews: http://www.creem.st-and.ac.uk/datamining/structure.html
b. Another one in Sweden: http://www.liu.se/utbildning/pabyggnad/F7MSM/courses?l=en
c. Stanford: https://statistics.stanford.edu/academics/ms-statistics-data-science
2. The lectures could have easily been recorded, this would have greatly enhanced the quality of the MSc. All you need is slides and a screen capture software with audio recording capability.
[Update: SOMAS now records the lectures in real time, and posts them on youtube. This has significantly improved course quality in my opinion, because it allows you watch an expert do the derivations on the board, and learn by copying/modeling that expert's approach to problem solving.]
3. The real value added in the MSc is the exercises, and the feedback after the exercises have been submitted. This is the only way that one learns new things in this course (apart from reading the lecture notes). The written exams are of course a crucial part of the program, but the solutions and one's own attempt are never released so one has only a limited opportunity to learn from one's mistakes in the exam. For about 2000 pounds a year, this is quite a bargain. Basically this is equivalent to hiring a statistician for 33 hours at 60 pounds an hour each year, with the big difference that you leave the table knowing much more than when you arrived.
4. Some ideas that were difficult for me:
- Expectation of a function of random variables was taught in the grad cert in 2011, but I needed it for the first time in 2014, when studying the EM algorithm. It would have been helpful to see a practical application early.
- The exponential distribution is a key distribution and needs much more study, esp. in connection with modeling survival. Perhaps more time should be spent studying distributions and their interrelationships.
- The derivation of full conditional distributions could have been tightly linked to DAGs, as is done in the Lunn et al book. It was only after I read the Lunn et al book that I really understood how to work out the full conditional distribution in any (within reason) given Bayesian model.
- I learnt how to compute eigenvalues and eigenvectors in the graduate certificate, but didn't use this knowledge until 2014, when I did Multivariate Analysis. I didn't even understand the relevance of eigenvalues etc. until I saw the discussion on Principal Components Analysis. A tighter linkage between mathematical concepts and their application in statistics would be useful.
- Similarly, Lagrangian multipliers became extremely useful when we started looking at PCA and Linear Discriminant Analysis; I saw them in 2011 and forgot all about them. There must be some way to show the applications of mathematical ideas in statistics. After much searching, I found this useful book that does part of the job.
5. The entire MSc program basically provides the technical background needed to understand major topics in statistics; there is not enough time to go into much detail. Each chapter in each course could have been a full course (e.g., the EM algorithm). I think that the real learning will not begin until I start to apply these ideas to new problems (as opposed to, say, using already known routines like linear mixed models). So, what I can say is that after four years of hard work, I know enough to actually start learning statistics. I don't feel like I really know anything; I just know the lay of the land.
6. The MSc is heavily dependent on R. Not having a python component to the course limits the student greatly, especially if they are going to go out there into the world as a ''data scientist''. The Enthought on-demand courses are a fantastic supplement to the MSc coursework. It would be a good idea to have a python course of that type in the MSc coursework as well.
7. One mistake I made from the perspective of exam-taking was not to spend enough time during the year using the hand-calculator (actually, I spent no time on this). In the exam, the difference between a distinction and an upper second can be the speed with which you can compute (correctly!) on a calculator. I am terrible at this, rarely even able to do simple calculations correctly on a hand-held (I'm talking about really basic operations), simply because I don't use calculators in real life; who does? I would have much preferred exams that test analytical ability rather than ability to do calculations quickly on a calculator. In the real world one uses computers to do calculations anyway. I was also hindered by the fact that I am half-blind (a side effect of kidney failure when I wa 20) and can't even see the hand-calculator's screen properly.
8. One peculiar aspect, and this permeated the MSc program, was the fairly antiquated instructions to students for using LaTeX etc. I think that statisticians should lead the way and use tools like Sweave and Knitr.
9. The textbook recommendations are out of date should be regularly revised. The best textbooks I found for each course that had exams associated with it:
Linear modelling: An Introduction to Generalized Linear Models, Dobson et al
Dobson et al is the best textbook I have ever read on generalized linear models, bar maybe McCullagh and Nelder. Dobson et al was a recommended book in the linear modeling course, a very good choice.
Bayesian Statistics: Lynch, Lunn et al, BDA3, Box and Tiao
Lynch is the best first book to read for Bayes (if you know calculus), and Lunn et al is very useful indeed, and beautifully written. It prepares you well for doing practical data analysis. Unfortunately, it's oriented towards WinBUGS, but one can translate the code easily to JAGS. In my opinion, WinBUGS was a great first attempt, but it should be retired now, because it is just so painful to use. People should go straight to JAGS (thanks to Martyn Plummer for doing just a fantastic job with JAGS) and then (or alternatively) Stan (thanks to Matt Hoffman, Bob Carpenter, Andrew Gelman and the Stan team for making it possible to use Bayes for really complex problems). You really need both JAGS and Stan in order to read and understand books, especially if you are just starting out.
I recommend reading Box and Tiao at the very end, to get a taste of (a) outstanding writing quality, and (b) what it was like to do Bayes in the pre-historic era (i.e., the 1970s).
Computational Inference: Statistical Computing with R, Rizzo
This book covers pretty much all of computational inference in a very user-friendly way,
Multivariate Analysis: Mathematical Tools for Applied Multivariate Analysis, By Carroll et al.
This book is very heavy going and not an after-five kind of book, it needs serious and slow study. I used it mostly as a reference book.
Medical Statistics (Survival Analysis): Regression Modeling Strategies by Harrell, and Dobson et al. I found the presentation of Survival Analysis in Harrell's book particularly helpful.
Concluding remarks
This MSc program is very valuable for someone willing to work hard on their own, with rather variable amounts of guidance from the instructors. It provides a lot of good-quality structure, and it allows you to check your understanding objectively by way of exams.
Summary of grades: You can see that I was starting to improve with experience!
YEAR 1
63% in Stats Lab
64% in Linear Modelling
YEAR 2
63% in Data Analysis
67% in Inference
YEAR 3
67% in Medical Statistics
70% in Dependent Data (Distinction)
73% in MSc dissertation (Distinction)
Overall grade: Pass with Merit.
Doing this MSc changed a lot of things for me professionally:
Teaching:
- I rewrote my lecture notes, abandoning the statistics textbook I had written in 2011. The Sheffield coursework played a huge role in helping me clean up my notes. I think these notes still need a lot of work, and I plan to work on them during my coming sabbatical.
-I started teaching undergrad Math as a prerequisite to my more technically oriented stats courses.
- I started teaching Bayesian statistics as a standard part of the graduate linguistics coursework. There doesn't seem to be much interest among most linguistics students in this stuff, but I do attract a very special type of student in these classes and that makes teaching more fun.
- I started teaching linear (mixed) modeling in a way aligns much more with standard presentations in the Sheffield MSc program.
- At least one of my students has taken advantage of Bayesian methods in their research, so it's starting to have an impact.
Research:
- One thing that became clear (if it wasn't obvious already) is that becoming a professional statistician or at least acquiring professional training in statistics is a necessary condition to doing analyses correctly, but it isn't a sufficient condition. Statisticians usually are unable to address concerns from people in specific areas of research because they have no domain knowledge. It seems that without domain knowledge, statistical knowledge is basically useless. One should not go to statisticians seeking "recommendations" on what to do in particular situations. Depending on which statistician you talk to, you can get a very variable answer. Coupled with knowledge of your research area and knowledge of statistical theory (which of course you have to acquire, just as you acquired your domain knowledge), you have to work out the answer to your particular problem.
- I have essentially abandoned null hypothesis significance testing and just use Bayesian methods. The linear modeling and Bayesian statistics plus computational inference courses were instrumental in making this transition possible. I still report p-values, but only because reviewers and editors of journals insist on them.
- I run high-powered studies whenever possible (e.g., it's not possible to run high power studies with aphasic populations, at least not at Potsdam). Everything else is a waste of time and money.
- I started posting all data and code online as soon as the associated paper is published.
-I spend a lot of time visualizing the data and checking model assumptions before settling on a model.
- I use bootstrapping a lot more to check whether my results hold up compared to more conventional methods.
- I try to replicate my results, and try to publish replications both of my own work and of others (much more difficult than I anticipated---people think replication is irrelevant and uninformative once someone has published a result with p less than 0.05.
- I can understand books like BDA3. This was not true in 2011. That was the biggest gain of putting myself through this thing; it made me literate enough to read technical introductions.
- I have started working on statistical problems and trying to publish methods papers. Two recent examples:
http://arxiv.org/abs/1506.06201
http://arxiv.org/abs/1506.04967
Subscribe to:
Comments (Atom)