We have just had a paper accepted in the journal Computational Brain and Behavior. This is part of a special issue that responds to the following paper on linear mixed models:
van Doorn, J., Aust, F., Haaf, J.M. et al. Bayes Factors for Mixed Models. Computational Brain and Behavior (2021). https://doi.org/10.1007/s42113-021-00113-2
There are quite a few papers in that special issue, all worth reading, but I especially liked the contribution by Singmann et al: Statistics in the Service of Science: Don't let the Tail Wag the Dog (https://psyarxiv.com/kxhfu/) They make some very good points in reaction to van Doorn et al's paper.
van Doorn, J., Aust, F., Haaf, J.M. et al. Bayes Factors for Mixed Models. Computational Brain and Behavior (2021). https://doi.org/10.1007/s42113-021-00113-2
There are quite a few papers in that special issue, all worth reading, but I especially liked the contribution by Singmann et al: Statistics in the Service of Science: Don't let the Tail Wag the Dog (https://psyarxiv.com/kxhfu/) They make some very good points in reaction to van Doorn et al's paper.
Our paper:
Shravan Vasishth, Himanshu Yadav, Daniel J. Schad, and Bruno Nicenboim. Sample size determination for Bayesian hierarchical models commonly used in psycholinguistics. Computational Brain and Behavior, 2021.
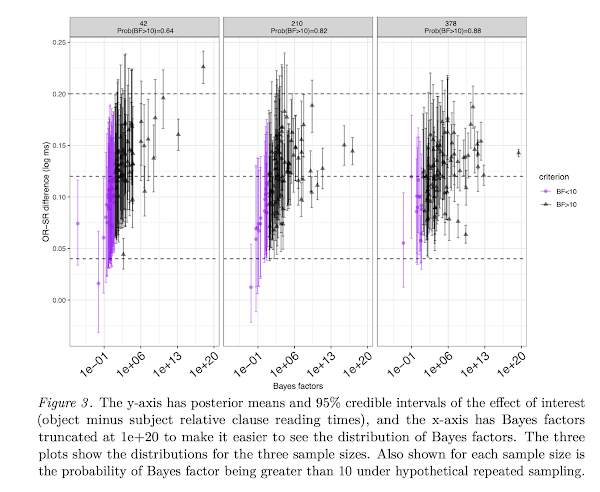
Abstract: We discuss an important issue that is not directly related to the main theses of the van Doorn et al. (2021) paper, but which frequently comes up when using Bayesian linear mixed models: how to determine sample size in advance of running a study when planning a Bayes factor analysis. We adapt a simulation-based method proposed by Wang and Gelfand (2002) for a Bayes-factor based design analysis, and demonstrate how relatively complex hierarchical models can be used to determine approximate sample sizes for planning experiments.
Code and data: https://osf.io/hjgrm/
pdf: here
Abstract: We discuss an important issue that is not directly related to the main theses of the van Doorn et al. (2021) paper, but which frequently comes up when using Bayesian linear mixed models: how to determine sample size in advance of running a study when planning a Bayes factor analysis. We adapt a simulation-based method proposed by Wang and Gelfand (2002) for a Bayes-factor based design analysis, and demonstrate how relatively complex hierarchical models can be used to determine approximate sample sizes for planning experiments.
Code and data: https://osf.io/hjgrm/
pdf: here